CISTO Scientists Adapt MaxEnt to Address the
"Big Data” Challenges of Climate Science

Left: Dr. John L. Schnase. NASA photo. Center: Cassin’s Sparrow (Peucaea cassinii). Photo by Bill Bouton; acquired from Wikimedia Commons. Right: Dr. Mark L. Carroll. NASA photo.
MAXENT
MaxEnt is an open-source, stand-alone Java application with a user-friendly interface that has become popular with biogeographers and ecologists for modeling and mapping the environmental niches and distributions of various species. MaxEnt — which refers to “maximum entropy” — and its machine learning (ML) algorithm derive distributions in geographic space that account for the maximum entropy, or statistical uncertainty, in the data.
MaxEnt was originally developed in 2002 by two colleagues from AT&T Labs in New York City — Steven Phillips, a computer science researcher, and Robert Schapire, an ML expert. The software addressed a need by the Center for Biodiversity and Conservation (CNC) at the American Museum of Natural History (AMNH) to use their large collection of observational datasets of species occurrences to model species distributions. The “black box” version of MaxEnt was hosted online by Princeton University for 10 years. Due to its popular use, an open-source version of the software was released in March 2017. Now maintained and hosted online by the CNC, users can find and download source code, datasets, and tutorials written in English, Spanish, and Russian and access the MaxEnt community’s discussion group.
BIG DATA CHALLENGES
“Big Data” is the term used to describe datasets that are so large and complex, delivered so fast and from such diverse sources that it is impractical for researchers to analyze using traditional methods. Climate change research has given rise to new technology requirements at the intersection of Big Data, ML, and high-performance computing (HPC). There are few places where this is more clearly seen than with studies that focus on Earth’s climate and its impact on species distribution and abundance. For nearly 20 years, the ecological modeling community’s tool of choice for this work has been MaxEnt. Few ML programs have been more widely used or more carefully studied.
Global climate models (GCMs) provide global representations of the climate system, make projections for hundreds of variables, and combine observations from an array of satellite, airborne, and in-situ sensors. These models can be configured to produce reanalyses that model the observed state of the global atmosphere in the past, such as the NASA Global Modeling and Assimilation Office’s (GMAO) Modern-Era Retrospective Analysis for Research and Applications, Version 2 (MERRA-2).
Today, there is increasing interest in using GCM outputs as predictors in ecological niche modeling. Researchers would like to take advantage of the growing number of climate datasets from an array of sensors, yet they are encountering difficulties with using such Big Data in MaxEnt. Despite its ease of use, MaxEnt has a limited capacity to analyze more than a small number of predictors and observations, because, like many ML systems, all the data it operates on must be stored in main memory. This is a challenge for researchers who would like to use GCM data, but need a way to screen large datasets for the best predictors. These key variables could then be used to create more accurate models that are not overly complex or require impractically long run times to compute.
Study Goals
Led by Emeritus Senior Computer Scientist Dr. John Schnase and Senior Scientist Dr. Mark Carroll, a team of researchers at the NASA Goddard Space Flight Center’s Computational and Information Sciences and Technology Office (CISTO) is working to overcome MaxEnt’s Big Data limitations by testing a more effective method of using large, high-dimensional datasets, like those produced by GCMs, that contain a large number of features or independent variables. The goal of this study is to create increasingly realistic environmental predictors within a reasonable amount of run time.
If successful, this project also has the potential to benefit the bioclimatic research community that increasingly relies on MaxEnt to understand how climate change impacts species distributions and survival. By streamlining the use of Big Data using MaxEnt, the increase in independent, climate-related research publications would add to the body of research assessed by the Intergovernmental Panel on Climate Change (IPCC).
Project Approach
The largest and most sophisticated GCMs are sometimes referred to as “IPCC-class” models because of the important role they play in the work of the IPCC. Such models produce petabyte-scale collections comprising hundreds of variables, a volume that vastly exceeds what is generally used in bioclimatic modeling today. Moreover, the direct outputs of IPCC-class GCMs are being transformed into derived data products on an unprecedented scale. As a result, variable selection and model tuning — crucial aspects of any species distribution modeling effort — have become complicated issues.
This study reflects the research team’s efforts to address one of these complex challenges: variable selection as an initial step in the modeling process. Their approach involves an external-memory (or out-of-core), Monte Carlo optimization that allows MaxEnt to operate on data stored in the files system rather than a computer’s main memory. The research team implemented repeated random sampling in a high-performance cloud computing setting on the NASA Center for Climate Simulation’s (NCCS) Advanced Data Analytics Platform (ADAPT) to obtain results.
Monte Carlo optimizations are a common way of finding approximate answers to problems that are solvable in principle but lack a practical means of solution. Out-of-core algorithms process datasets that are too large to fit into a computer’s main memory, which is why such optimizations are currently a major focus of research in the ML community. The NASA research team’s solution brought these two concepts together in an ensemble approach that employed many parallel MaxEnt runs, each drawing on a small random subset of environmental variables stored in the file system. These runs converged on a global estimate of the top contributing subset of variables in the larger collection. Once those top variables were selected, those variables were then run in MaxEnt.
To better understand how climate and habitat changes influence species distributions, the CISTO team conducted MaxEnt runs to correlate an array of georeferenced, observational data of particular species occurrences — in this case, Cassin’s Sparrow (Peucaea cassinii) — with realistic climate models using the out-of-core Monte Carlo method implemented in ADAPT.
Cassin’s Sparrow was used as the target species for development testing. This sparrow is an elusive, dry grassland-adapted bird that typically lives and breeds in the central and southwestern United States, yet is nomadic and sometimes found far out of its normal range — as far north as Canada and on both coasts. Since desert-adapted birds are particularly vulnerable to climate change, this species is ideally suited for this study.
After overlapping observations were removed from 1,865 records downloaded from the open-access Global Biodiversity Information Facility (GBIF) database, 609 observations of species occurrences from within a 16-kilometer buffer were used. For predictors, the team used Worldclim’s standard bioclimatic (bioclim) environmental variables at a resolution of 5.0 arc-minutes throughout. These predictor layers were clipped to the coverage area of the observational data, reprojected, and formatted for use with MaxEnt using the Geospatial Data Abstraction Library Version 3.0 software.

Above: The 19 standard bioclim environmental variables used in this study.
During the Monte Carlo sampling runs, randomly selected environmental variables were iteratively tested for their predictive capacity and assigned a relative weight for a given cell in the studied landscape, in terms of habitat suitability to Cassin’s Sparrow. The current study focused on stochastic down-selection from a full variable set as a preliminary screening step to be refined prior to final model construction.
Since six or fewer predictors generally predominated in the environmental niche models for this study, the team used MaxEnt in two different ways to find the six most influential range — and niche — defining bioclim variables for Cassin’s Sparrow. In the first experiment, they developed a baseline model using the stand-alone MaxEnt program in the traditional way. In the second experiment, the team used the Monte Carlo approach in ensemble runs comprising 100 “sprints,” with each sprint consisting of ten MaxEnt runs operating on a small random subset of 19 bioclim predictors read from the filesystem.
A tally table was used to maintain a count of the number of times a variable was used in a run along with a cumulative sum of the variable’s permutation importance. The tally table thus provided the information needed to determine the average permutation importance of a predictor at any point along the way. This resulted in an evolving progression of models that converged on a stable assemblage of top six predictors over the course of an ensemble. Collectively, the team’s Monte Carlo ensembles resulted in more than 2,000 MaxEnt runs.
A distinct pattern of progression toward a stable subset of key variables was observed in the Monte Carlo ensembles: the top three contributory variables among the top six were selected early in the sprint runs, and measures of relative explanatory power fluctuated within a narrow range, with little variation during the course of the selection process.
Results
The team studied several attributes of the six-variable final models to ascertain how the Monte Carlo algorithm performed. The predictive distribution maps produced by the models were evaluated for reasonableness based on first-hand knowledge of the species, its habitat preferences, known range from observational records, and statistical analysis. Single-processor run times were recorded to help identify opportunities for multiprocessor parallelization.
Early results indicated that the Monte Carlo method used in this study reliably selected a suitable subset of the original predictors, which could then be explored in more detailed ways and further refined prior to final model construction. Since each model run in the Monte Carlo screening process is independent and uses a set number of variables, the method is completely parallelizable, independent of the innate scaling properties of MaxEnt, and useful to implement as an external memory algorithm. If proven effective, such an approach could contribute to the ecological modeling process when there is a need to preselect a small set of predictors in a pool comprising a potentially very large number of predictors. This could lead to greater use of climate model outputs by the ecological research community and aid the search for viable predictors when the selection of variables through ecological reasoning is not apparent.
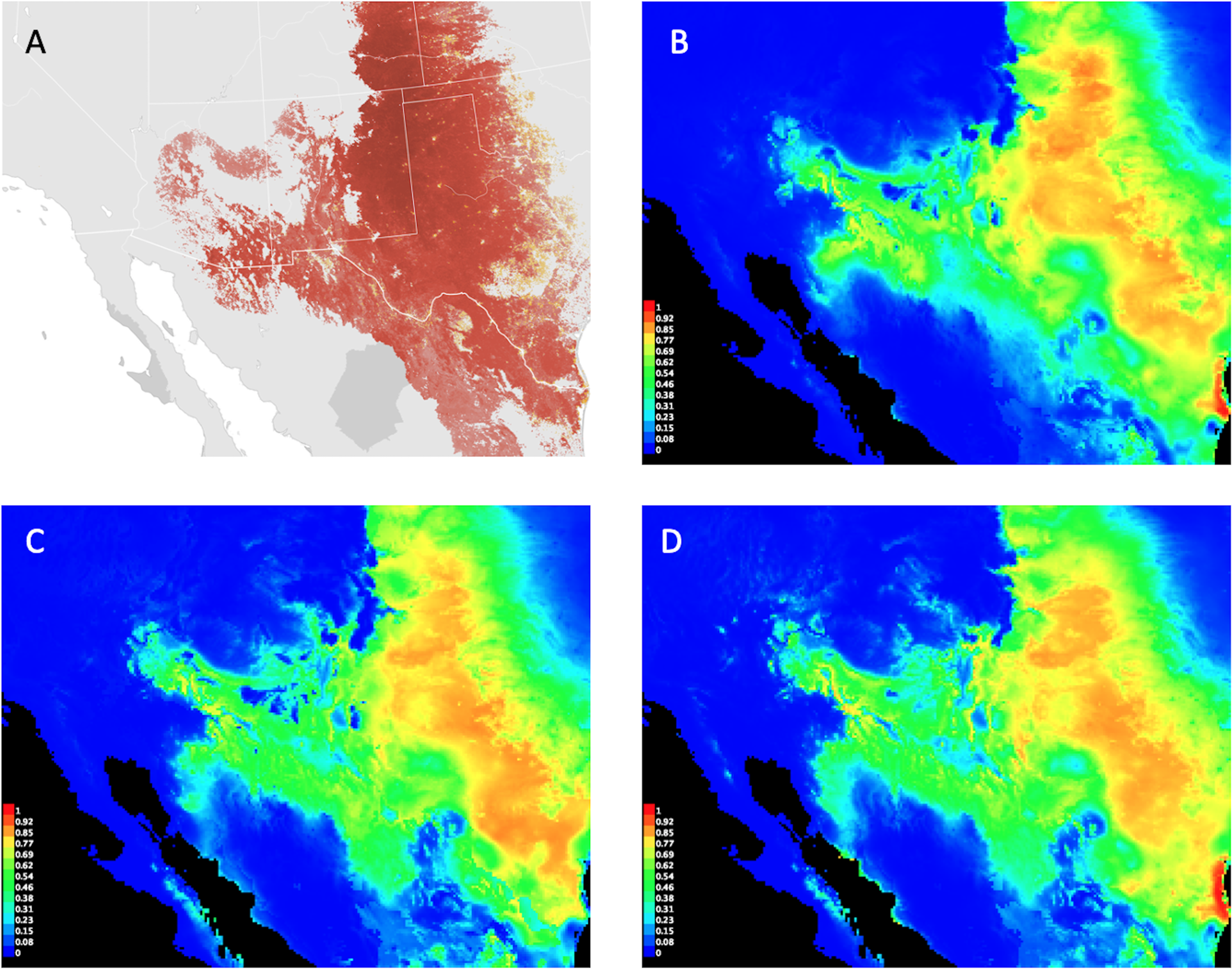
(A) Cassin’s Sparrow range map based on observational records from 2016; compared to the predicted habitat suitability distributions from (B) MaxEnt baseline, (C) Monte Carlo Ensemble #1, and (D) Monte Carlo Ensemble #2. Images (B)–(D) created by the research team show MaxEnt logistic output, an estimated probability of presence between 0 and 1 with warmer colors indicating better-predicted conditions for the species. Image (A) provided by eBird (www.ebird.org) with permission from the Cornell Lab of Ornithology.
SAVING TIME WITH EMBARRASSINGLY PARALLEL SUBTASKS
Depending on the numbers of variables used, baseline MaxEnt runs took from 18 minutes to over two hours, with some traditional runs taking nearly 30 hours to complete. However, since each of the Monte Carlo MaxEnt runs was entirely independent from all other runs in the ensemble, “embarrassingly parallel” workloads on multiprocessor implementations of the method used are possible. For example, using high-performance cloud computing, a 1,000-run ensemble used to select the most influential variables as an initial step in the modeling process could conceivably take only as long as a single MaxEnt run. In most cases, that would be about one minute! Clearly, this study achieved its goal to create increasingly realistic environmental predictors within a reasonable amount of run time.
COMPUTE USED
The team implemented this project in a 100-core testbed using ADAPT, a managed virtual machine (VM) environment most closely resembling a platform-as-a-service (PaaS) cloud. ADAPT features over 300 physical hypervisors that host one or more VMs, each having access to multiple shared, centralized data repositories. The hypervisor hardware consists of 2.2 GHz 24-core Intel Xeon Broadwell E5-2650 v4 processors with 256 gigabytes of memory. The testbed consists of a dedicated set of ten 10-core Debian Linux 9 Stretch VMs. To develop the software components of this project, the team used MaxEnt version 3.4.1; shell scripts; R version 4.0.1; and ENMeval version 0.3.1.
In evaluating the impact of this study, Carroll observed, “this Big Data approach to selecting variables for machine learning has potential applications to comprehensive, high-dimensional, multi-decade data such as global climate reanalysis and data from hyperspectral remote sensing of Earth.”
“Initial results show that this approach can be used effectively on larger data collections,” Schnase further remarked. “It is capable of achieving nearly real-time performance in a multiprocessor environment and can be used to identify a reasonable subset of top environmental predictors in our test collections. While still at an early stage of development, the study provides insight into the potential opportunities afforded by Monte Carlo techniques, high-performance cloud computing, and process automation in addressing some of the Big Data challenges facing the ecological research community.”
Schnase’s longitudinal research to understand Cassin's Sparrow led to the application of avian energetics modeling using computers. This study should help continue that work, addressing research questions relating to key climatic influences on this nomadic bird’s distribution and effectively incorporating GCMs using MaxEnt.

Two-dimensional projection of a hyperspectral data cube.
Image by Dr. Nicholas M. Short, Sr.
Next Steps
The CISTO research team will next focus on testing a parallel, high-performance version of the Monte Carlo approach in the ADAPT science cloud using the Global Modeling and Assimilation Office’s (GMAO) Goddard Earth Observing System, Version 5 (GEOS-5) climate modeling system. Researchers plan to extend the randomly determined selection of feature class and regularization multiplier and make improvements to the algorithm, such as developing better measures of variable importance. Schnase added, “we look forward to evaluating the effectiveness of the new method by using it to research climate change influences on the distribution of Cassin’s Sparrow.”
Related Links
- Accelerating Science with AI and Machine Learning, NCCS Highlight, 12/18/20.
- Cassin's Sparrow: One Small Bird's Message to a World Whose Past, Present, and Future We Share.
- Schnase, J.L., M. Carroll, R. Gill, G.S. Tamkin, J. Li, S. Strong, T. Maxwell, M.E. Aronne, and C.S. Spradlin, 2021: Toward a Monte Carlo Approach to Selecting Climate Variables in MaxEnt. PLOS ONE 16(3), doi:10.1371/j.pone.0237208.
- Schnase, J.L., D.Q. Duffy, G.S. Tamkin, D. Nadeau, J.H. Thompson, et al., 2017: MERRA Analytic Services: Meeting the Big Data Challenges of Climate Science Through Cloud-Enabled Climate Analytics-As-A-Service. Comput Environ Urban Syst, 61, doi:10.1016/j.compenvurbsys.2013.12.003.
- Salas, E.A.L., V.A. Seamster, K.G. Boykin, N.M. Harings, K.W. Dixon, 2017, Department of Fish, Wildlife and Conservation Ecology, New Mexico State University, Las Cruces, New Mexico 88003, USA. Modeling the Impacts of Climate Change on Species of Concern (birds) in South Central U.S. Based on Bioclimatic Variables. AIMS Environ. Sci., 4, doi:10.3934/environsci.2017.2.358.
Sean Keefe, NASA Goddard Space Flight Center


