FYI: NCCS NUGGETS
JupyterHub Disconnects
Issue to be resolved:
Frequent disconnects from JupyterHub with one of these messages:
- “Server unavailable or unreachable. Would you like to restart it?”
- “Kernel restarting: The kernel appears to have died. It will restart automatically.”
Solution:
“Server unavailable or unreachable. Would you like to restart it?”
This message is exclusively related to intermittent network/connectivity issues between your client and the JupyterHub server. You may simply refresh the page to connect to an active JupyterHub session or start a new session if the session timeout has been reached. If you are onsite at GSFC and are regularly experiencing network issues, please contact the Enterprise Service Desk (ESD) to report those network problems:
https://esd.nasa.gov/esdportal
These prompts may also occur if your authentication through NASA’s Access Launchpad expires. Again, simply refresh the page or start a new session. Launchpad timeouts are outside of NCCS control and most frequently occur during JupyterHub sessions that last longer than 8 or 12 hours.
“Kernel restarting: The kernel appears to have died. It will restart automatically.”
This could also be caused by intermittent network connectivity, but usually it indicates either an out of memory issue or a software fault. You may need to either request a new session with more memory or investigate reducing the memory you use. You can also check your /home directory for logs. For example, “jhub-XXXX.out” on Discover or “slurm-XXXX.out” on ADAPT. These may offer a clue as to why the kernel is dying.
Please also consider submitting a batch job via Slurm for any long-running work. See the Slurm documentation below:
Discover: https://www.nccs.nasa.gov/nccs-users/instructional/using-slurm
ADAPT: https://www.nccs.nasa.gov/nccs-users/instructional/adapt-instructional/s...
Date: 02/10/26
Issue to be resolved: Frequent disconnects from JupyterHub with one of these messages.
Conda Environments
Issue to be resolved:
Creating a Conda environment or installing a package causes an indefinite hang.
Solution:
There are a few steps required to mitigate this issue. It is caused by Conda attempting to search/install from the “defaults” channel.
1) Ensure you are using the miniforge module, and NOT anaconda.
2) Also ensure your .condarc file (in your home directory) does not contain the “defaults” entry under the channels section by running the following commands:
> module load miniforge
> conda config --add channels conda-forge
> conda config --remove channels defaults
No further steps are needed if you’re using conda create.
However, conda install has a caveat. When invoked, it will try to search all channels that correspond to packages in the environment. This means that for environments already containing licensed Anaconda packages, conda install will attempt to read from “defaults” again. This will fail as expected. You can simply run the exact same command again and conda install will drop “defaults.”
NOTE: For conda create and conda install — If necessary, you can always specify the channel with -c <channel> and --override-channels.
For example:
> conda install numpy -c conda-forge --override-channels
> conda install cuda -c nvidia --override-channels
3) If creating the environment using a YAML file, (via “conda env create -f <filename>”) you can add the following to the environment file:
channels:
If you need any other channels, you can add them as well (e.g. “nvidia”). Using “nodefaults” will instruct Conda not to use the default Anaconda packages, which require a license.
For more information: See the Conda documentation
Date: 09/24/25
Issue to be resolved: Creating a Conda environment or installing a package causes an indefinite hang.
Large Resource Requests
Issue to be resolved:
My job requires more time or resources than are currently allowed.
Solution:
Email the NCCS at support@nccs.nasa.gov and detail the resources constraint(s) that prevent your job from running. Provide an assessment of the resources, compute (CPU or GPU), RAM, and/or time, that would be required to complete your work. We will likely reach out to discuss your use case so that we can determine how best to meet your needs.
Note, the NCCS employs constraints on jobs for many reasons, key among them:
1. To prevent runaway jobs from consuming all available resources until systems staff can kill the offending job
2.To prevent users from running days long jobs with no restart capability that fail due to an external cause, resulting in the loss of days of compute and requiring the job to be restarted from the beginning
3. To prevent a large set of jobs from dominating the queue, thereby preventing other users from accessing the resources for days at a time
For further information:
Slurm Best Practices on Discover
Date: 07/16/25
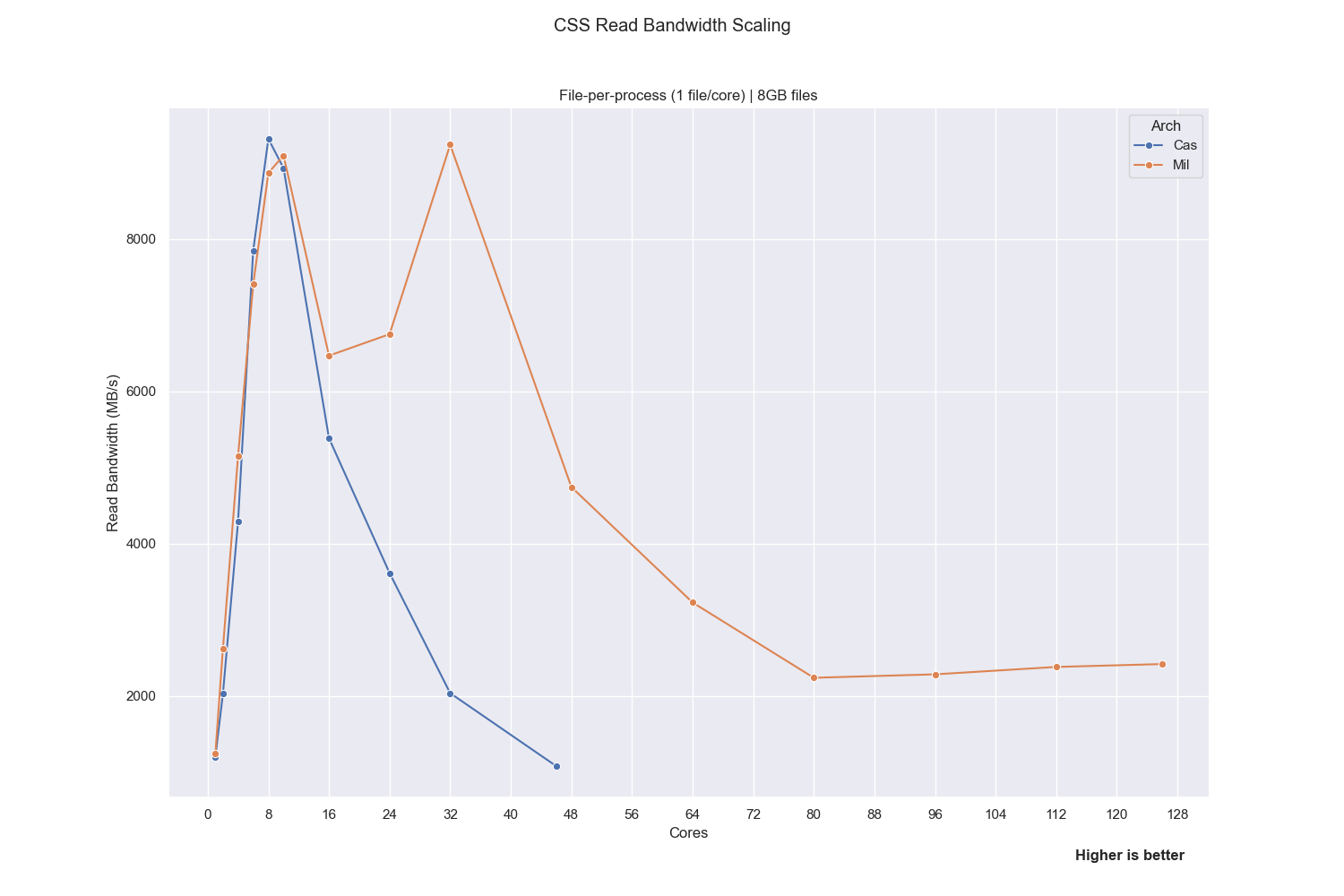
Issue to be resolved: Reading from CSS in parallel is very slow on Discover.
CSS Read Performance
Issue to be resolved:
Reading from CSS in parallel is very slow on Discover.
The NCCS doesn't currently run an Anaconda license server and does not plan to do so. Note, we are still working through this issue but we wanted to get some information out to our user community as soon as possible.
Solution:
To maintain higher bandwidth, use fewer than 12 cores/processes when performing read operations. CSS is a NFS filesystem that does not support robust parallelization. Internal testing has shown that reading from CSS with more than 12 cores can degrade bandwidth significantly.
This degradation can be exacerbated if multiple users read in parallel at the same time.
Date: 07/16/25
Issue to be resolved: Reading from CSS in parallel is very slow on Discover.
Anaconda License Issue
Issue to be resolved:
Anaconda is now requiring a paid license:
https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.an...
as part of their Terms of Service:
https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Flegal....
The NCCS doesn't currently run an Anaconda license server and does not plan to do so. Note, we are still working through this issue but we wanted to get some information out to our user community as soon as possible.
Solution:
1. Short term: Use Miniconda:
https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdocs.a...
or another opensource replacement and ensure that you are not including any default packages that would come from Anaconda
Use conda list --show-channel-urls and check the last column for the channel
Long term, move to Miniforge:
https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fgithub...
2. ADAPT: It may be possible to purchase Anaconda licenses for $199/user through NEST and use it on your ADAPT systems. We'll provide more information when we know more.
Date: 08/29/24
Issue to be resolved: Anaconda is now requiring a paid license.
Discover Temporary Storage
Issue to be resolved: Where is the best place to write temporary (scratch) data on Discover?
Solution: Discover has multiple scratch filesystems: $LOCAL_TMPDIR, $TMPDIR, $TSE_TMPDIR.
Each of these environment variables are created during login sessions for interactive use as well as each individual SLURM batch job.
$LOCAL_TMPDIR is node specific so any data stored there can only be accessed from the particular node where it is placed. $TMPDIR is accessible from any node and has a default quota of 5TB and 300k inodes.
To increase the I/O performance of jobs that read and write temporary files, there is a fast scratch filesystem using NVMe (flash) technology.
Access this storage by using $TSE_TMPDIR where you might otherwise use $TMPDIR. Each user has a default quota of 250 GB and 200,000 inodes on $TSE_TMPDIR. If you have a job that needs more than the default allocation, please email us at support@nccs.nasa.gov.
*Important*
* The data stored in the TMPDIR environments are not backed up and are deleted at the end of your job or login session because these resources are very limited.
* Do not specify $ARCHIVE for temporary storage. It is not part of the local Discover filesystems. It is a tape library mounted over the network and most users do not have access because it is a deprecated service.
Additional information on Discover Filesystem and Storage can be found on the NCCS Web-site at the following URL:
https://www.nccs.nasa.gov/nccs-users/instructional/using-discover/file-s...
Date: 04/11/24
Issue to be resolved: Where is the best place to write temporary (scratch) data on Discover?
Mac SSH Login Errors
Issue to be resolved: MacOS 13.4 users may be seeing the following error when they attempt to login to NCCS systems (Discover, Dirac, ADAPT):
Infinite reexec detected; aborting
The error you are seeing is a newly found bug that Apple introduced in MacOS 13.4 (this is something specific to how MacOS is handling PKCS11/PIV and interaction with the ProxyCommand/ProxyJump options that are used for access through a bastion node like login.nccs.nasa.gov).
Solution: To work around this issue, please create a file with the following aliases:
$ cat $HOME/.ssh/ssh_alias_workaround
# create aliases to use ssh-apple-pkcs11 directly to work around # "Infinite reexec detected" error alias ssh=/usr/libexec/ssh-apple-pkcs11 alias scp="scp -S /usr/libexec/ssh-apple-pkcs11"
alias sftp="sftp -S /usr/libexec/ssh-apple-pkcs11"
alias rsync="rsync -e /usr/libexec/ssh-apple-pkcs11"
You can then "source" this file directly in your shell on your MacOS system or add that source into your shell init (.bashrc/.cshrc/.zshrc):
source $HOME/.ssh/ssh_alias_workaround
You can check to see if the aliases are present in the shell after source by just running the "alias" command without any option.
You should then be able to test your scp/sftp/rsync/ssh command again which should now work without having to change your originally working $HOME/.ssh/config.
This workaround may be needed for some time, we don't currently have an expected date for a fix from Apple (or any sort of indication on how they plan to address the issue).
For more information: If you try the above and continue to have problems, please email support@nccs.nasa.gov
Date: 06/07/23
Issue to be resolved: MacOS 13.4 users may be seeing the following error when they attempt to login to NCCS systems (Discover, Dirac, ADAPT).
Discover Fast Temporary Storage
Issue to be resolved: Slow Discover jobs due to $TMPDIR I/O performance
Solution: For jobs that read and write temporary files, there is a fast scratch filesystem using NVMe (flash) technology. Access this storage by using $TSE_TMPDIR where you would currently use $TMPDIR.
Each user has a default allocation of 250 GB and 200,000 inodes. This data is not backed up and is deleted at the end of your job because the resource is very limited. If you have a job that needs more than the default allocation, email support@nccs.nasa.gov so that we can discuss your job requirements.
For more information on available Discover storage:
https://www.nccs.nasa.gov/nccs-users/instructional/using-discover/file-s...
Date: 10/31/22
Issue to be resolved: Slow Discover jobs due to $TMPDIR I/O performance.
Specifying multiple CPUs types for Discover jobs
Issue to be resolved: Lowering Discover wait times.
Solution: Modify your Slurm submission scripts to use allow Slurm to send your job to Haswell or Cascade Lake cores.
Additional motivation: Discover's Haswell nodes will be decommissioned before the end of the calendar year so this would be a great time to make sure your code runs on the Cascade Lake nodes.
The NCCS recently changed the default cores to be selected by Slurm from "Haswell" to "Cascade Lake and Haswell", with Cascade Lake being the preferred choice. This could impact your jobs in two ways:
1. If your Slurm submission script specifies ' --constraint=hasw ', then your jobs will not be sent to the newer nodes.
2. If your Slurm submission script specifies ' --constraint=cas ', then your jobs will now be competing with more jobs for a finite resource.
Using ' --constaint="hasw|cas" ' will allow Slurm to schedule your job efficiently. (Enclosing the string in double quotes is required to ensure that the shell does not interpret the "|" symbol as a pipe character.) Note that Slurm treats these values as order-independent. If you have a strong preference for Cascade Lake, you can also specify ' --prefer=cas ' (introduced in v22.05).
Please note the following:
1. If your code can also run on the Skylake nodes, use ' -- constraint="hasw|sky|cas" .'
2. If code that has been running successfully on Haswell nodes generates an error on the Cascade Lake nodes, please submit a ticket to support@nccs.nasa.gov.
Additional information on Slurm best practices can be found here.
Date: 09/15/22
Issue to be resolved: Lowering Discover wait times.
Prism A100 GPUs
Issue to be resolved: Need access to faster GPU nodes with more RAM.
Solution: Submit your jobs to the "DGX" QoS on Prism by specifying "-p dgx". This gives you access to up to 8 NVIDIA A100 GPUs with:
* 8x NVIDIA A100 GPUs with 40 GB of VRAM and NVLink
* Dual AMD EPYC Rome 7742 CPUs; 64 cores each at 2.25GHz
* 1 TB RAM
* Dual 25Gb Ethernet network interfaces
* Dual 100Gb HDR100 Infiniband high speed network interfaces
* 14 TB RAID protected NVMe drives, mounted as /lscratch
All Prism users have access to the dgx QoS. Note, there are 22 nodes with NVIDIA V100 GPUs (4 GPUs each) so they will be more readily available. If you are using the A100s for smaller jobs, you may be asked to move to the V100s if a larger job needs these resources.
Example, to request an interactive session with 2 x A100 GPUs:
salloc -G2 -p dgx
For more information:
https://www.nccs.nasa.gov/systems/ADAPT/Prism
Date: 08/22/22
Issue to be resolved: Need access to faster GPU nodes with more RAM.
Discover job wait times
Issue to be resolved: Long waits for small jobs to start on Discover.
Solution: Check your job summary report in your job's standard output/error file (goes to slurm-
Example below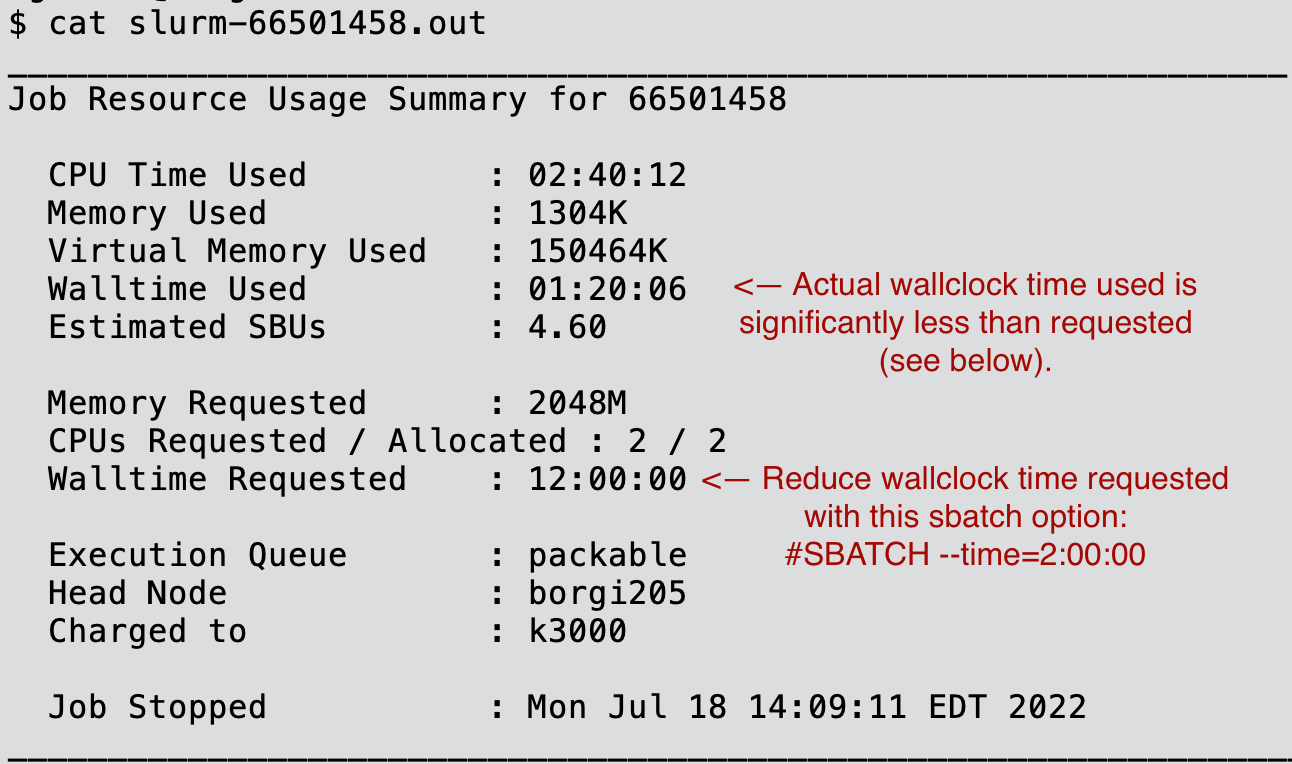
Additional information on Slurm best practices can be found here.
Date: 07/19/22
Issue to be resolved: Long waits for small jobs to start on Discover.
User Directory permissions
Issue to be resolved: Potential loss of data through world write directories as they allow other users to unintentionally or maliciously delete data you have in world writeable directories.
Solution:
1. Run these command to determine if you have any world write directories:cd $HOME
find ${HOME}/ -xdev -perm -o=w ! -type l -ls >& home_world_write_files.txtcd $NOBACKUP
find ${NOBACKUP}/ -xdev -perm -o=w ! -type l -ls >& nobackup_world_write_files.txt
If you own project nobackup spaces, you should do the same there.
2. If the above commands return any world writeable directories, change them with this command:
chmod o-w dirname
Where you substitute dirname with the actual directory name.
3. If you find you have a lot of world write directories in your home or nobackup directory, check your umask with this command:
umask
If it isn't "77", which is the default, we highly recommend that you change it with this command:
umask 77
to protect your data from unintentional or malicious deletion by processes run by other Discover users.
Additional information about Linux permissions can be found here.
Date: 02/15/22
Issue to be resolved: Potential loss of data through world write directories as they allow other users to unintentionally or maliciously delete data you have in world writeable directories.


